Research Highlights
(2026) Structure of SHOC2-KRAS-PP1C complex reveals RAS isoform-specific determinants and insights into targeting complex assembly by RAS inhibitors. Nat Commun. 10.1038/s41467-026-68319-1
(2025) The structural basis for the selective antagonism of soluble TNF-alpha by shark variable new antigen receptors. Nat Commun. 10.1038/s41467-025-66967-3
(2025) Structural basis for LZTR1 recognition of RAS GTPases for degradation. Science. 389, 1112-1117
(2025) Molecular glues that facilitate RAS binding to PI3Kα promote glucose uptake without insulin.. Science. 389, 402-408
(2025) CARD domains mediate anti-phage defence in bacterial gasdermin systems. Nature. 10.1038/s41586-024-08498-3
(2025) Structural insights into isoform-specific RAS-PI3Kα interactions and the role of RAS in PI3Kα activation.. Nat Commun. 16, 525
(2025) Dynamic allostery in the peptide/MHC complex enables TCR neoantigen selectivity. Nat Commun. 16, 849
(2025) Molecular basis of hemoglobin binding and heme removal in Corynebacterium diphtheriae. Proc Natl Acad Sci U S A. 122, e2411833122

(2024) Structural, biophysical, and biochemical insights into C-S bond cleavage by dimethylsulfone monooxygenase. Proc Natl Acad Sci U S A. 121, e2401858121

(2024) Preclinical proof of principle for orally delivered Th17 antagonist miniproteins. Cell. 187, 4305-4317.e18

(2024) Binding and sensing diverse small molecules using shape-complementary pseudocycles. Science. 385, 276-282
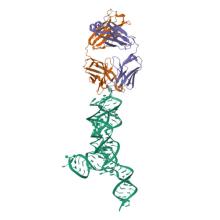
(2024) Structural basis for promiscuity in ligand recognition by yjdF riboswitch. Cell Discov. 10, 37
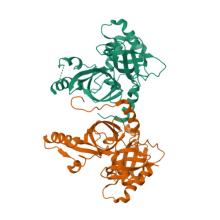
(2024) Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes. Nat Commun. 15, 2064

(2024) Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography. J Chem Inf Model. 64, 1704-1718
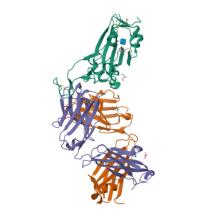
(2024) Diverse array of neutralizing antibodies elicited upon Spike Ferritin Nanoparticle vaccination in rhesus macaques. Nat Commun. 15, 200
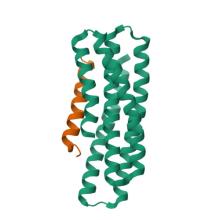
(2023) De novo design of high-affinity binders of bioactive helical peptides. Nature. 10.1038/s41586-023-06953-1
(2023) Engineered Antigen-Binding Fragments for Enhanced Crystallization of Antibody:Antigen Complexes. Protein Sci. 10.1002/pro.4824
(2023) Low complexity domains of the nucleocapsid protein of SARS-CoV-2 form amyloid fibrils. Nat Commun. 14, 2379
(2023) A Fluorescence Polarization Assay for Macrodomains Facilitates the Identification of Potent Inhibitors of the SARS-CoV-2 Macrodomain. ACS Chem Biol. 18, 1200-1207
(2023) Crystal structure of a highly conserved enteroviral 5' cloverleaf RNA replication element. Nat Commun. 14, 1955
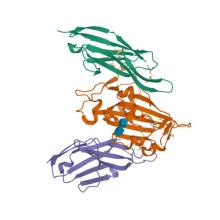
(2023) Shark nanobodies with potent SARS-CoV-2 neutralizing activity and broad sarbecovirus reactivity. Nat Commun. 14, 580
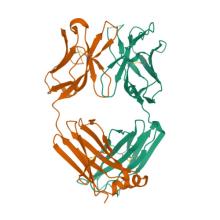
(2023) Hierarchical sequence-affinity landscapes shape the evolution of breadth in an anti-influenza receptor binding site antibody. Elife. 10.7554/eLife.83628
(2022) A unifying Bayesian framework for merging X-ray diffraction data. Nat Commun. 13, 7764
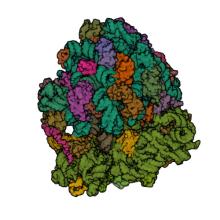
(2022) Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes. Nat Chem. 10.1038/s41557-022-01073-1
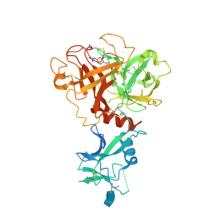
(2022) Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation. Nat Chem Biol. 10.1038/s41589-022-01059-7
(2022) An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses. Sci Transl Med. 10.1126/scitranslmed.abn6859